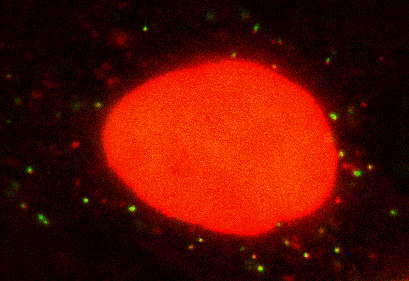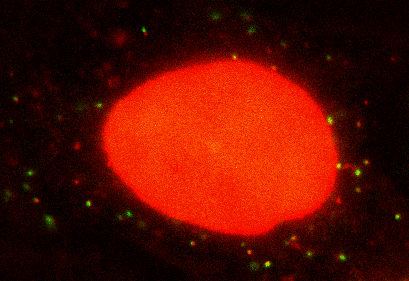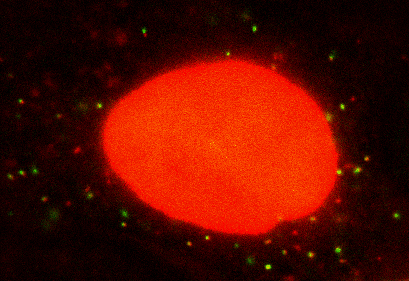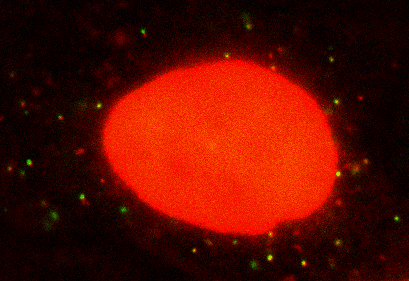
Seeing is believing: My lab is interested in visualizing and quantifying biological events as they happen in situ at real time. We use single molecule imaging and spectroscopy technology in live cells, combined with theoretical modeling to extract dynamic biological information. Currently, the lab is working on how cells regulate gene expression. Gene expression has to be tightly controlled both temporally and spatially for an organism to survive and prosper. Dysfunction in local regulation of gene expression is one of the most common sources of developmental and neurodegenerative disease. My laboratory focuses on this fundamental process by visualizing and manipulating the birth, trafficking, translation and decays of single RNA molecules in live cells and tissue. Recently we have developed a method called single molecule imaging of nascent peptides (SINAPS) to visualize translation of single mRNAs. With SINAPS, we can answer biological questions that are intractable before, such as the location of translation, the translation speed of ribosome in cells, the initiation and release rate of ribosome, the relationship between splicing, decay and translation, the effect of untranslated region on translation, etc. We are also interested in local translation in neuronal, such as the effect of synaptic stimulation and dendritic translation, local translation in the axonal growth cone. Besides purely observation, my lab is also focused on developing optic method to manipulate gene expression in live cells. Eventually, we hope to turn on or off expression of particular gene anywhere in a cell.
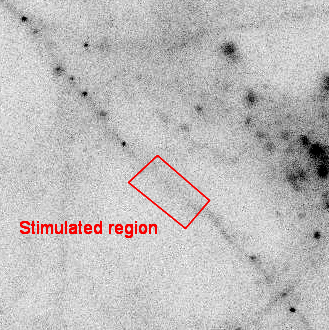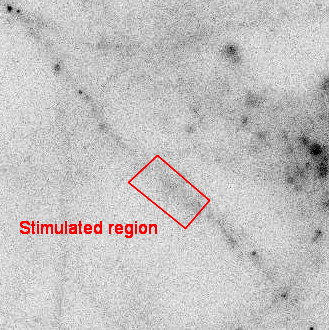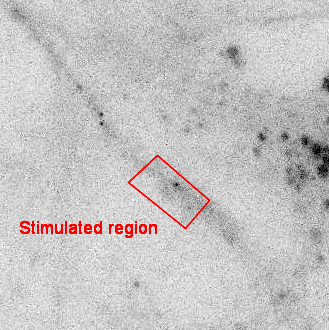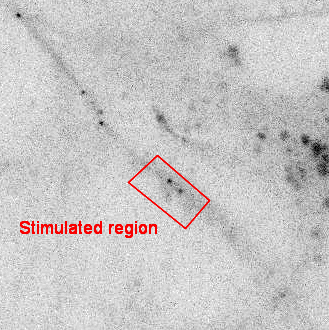
SINAPS
Bin Wu, Eliscovich C, Yoon Y and Singer R.H., Translation dynamics of single mRNAs in live cells and neurons, Science, Vol 352, Issue 6292, 2016, pp. 1430-1435, DOI:10.1126/science.aaf1084